Discovery of patterns (motifs) in biological sequences
Coordinator/contact:
This email address is being protected from spambots. You need JavaScript enabled to view it.
This email address is being protected from spambots. You need JavaScript enabled to view it.
Introduction
The Motif Discovery Problem (MDP) is an important biological problem whose main objective is to discover over-represented short patterns, named motifs, in a set of DNA sequences that may have some biological significance. More concretely, a motif is defined as a nucleic-acid sequence pattern that has some biological relevance, such as being DNA binding sites for a protein. Motifs are mixed with a large amount of biological information in DNA sequences; therefore, discovering them is not a trivial task. MDP tries to solve optimally the problem of finding motifs, applied to the specific task of discovering novel Transcription Factor Binding Sites (TFBS) in DNA sequences. Tools for the assessment of similarity and the identification of related sequences are among the most important tools in bioinformatics as they may serve for the functional categorization of novel genes or proteins of unknown function. To discover motifs with biological significance we must fulfil specific objectives while satisfying a variety of constraints. Thus, we proceed proposing a multiobjective formulation adequately adapted according to the real-world biological requirements, defining three conflicting objectives to be maximized: motif length, support, and similarity.
Motif Discovery Problem Formulation
Given a set of sequences S = { S_i | i = 1,2,...,D } of nucleotides defined on the alphabet B = { A,C,G,T }. S_i = { S_i^j | j = 1,2,...,w_i } is a sequence of nucleotides, where w_i is the sequence width. The set of all the subsequences contained in S is {s_i^{j_i} | i = 1,2,...,D, j_i = 1,2,...,w_i - l + 1 }, where j_i is the binding site of a possible motif instance s_i^j on sequence S_i, and l is the motif length, the first objective to be maximized. To obtain the values of the other two objectives we have to build the Position Indicator Matrix (PIM) A = { A_i | i = 1,2,...,D } of the motif, where A_i = { A_i^j | j = 1,2,...,w_i } is the indicator row vector with respect to a sequence S_i. A_i^j is 1 if the position j in S_i is a binding site, and 0 otherwise. We refer to the number of motif instances as |A| = \sum_{i=1}^D{\sum_{j=1}^{w_i}{A_i^j}}. We also require to find the consensus motif, which is a string abstraction of the motif instances. In this work we consider a single motif instance per sequence. Only those sequences that achieve a motif instance of certain quality with respect to the consensus motif were taken into account when we perform the final motif. This is indicated by the second objective, the support.
Furthermore, S(A) = { S(A)_1,S(A)_2,...,S(A)_{|A|}} is a set of |A| motif instances, where S(A)_i = S(A)_i^1S(A)_i^2...S(A)_i^l is the ith motif instance in |A|. S(A) can also be expanded as (S(A)^1,S(A)^2,...,S(A)^l), where S(A)^j = S(A)_1^jS(A)_2^j...S(A)_{|A|}^j is the list of nucleotides on the jth position in the motif instances.
Then, we build the Position Count Matrix (PCM) N(A) with the numbers of different nucleotide bases on each position of the candidate motifs (A) which have passed the threshold marked by the support. N(A) = { N(A)^1,N(A)^2,...,N(A)^l }, and N(A)^j = { N(A)_b^j | b \in{B} }, where N(A)_b^j = |{ S(A)_i^j | S(A)_i^j = b }|. The dominant nucleotides of each position are normalized in the Position Frequency Matrix (PFM) N = \frac{N(A)}{|A|}. Finally, we calculate the third objective, the similarity, averaging all the dominance values of each PFM column, as is indicated in the following expression:
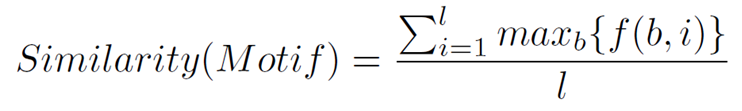
where f(b,i) is the score of nucleotide b in column i in the PFM and max_b{f(b,i)} is the dominance value of the dominant nucleotide in column i.
To guide the pattern search to solutions that have biological relevance, we have incorporated several constraints that should be satisfied by each solution. In motif discovery, motifs are usually very short, so that, finding long motifs means to lose a great computational time. To avoid this, we have restricted the motif length to the range [6,64], where the minimum is 6 and the maximum is 64. In the second objective we have also set a minimum support value of 2 for the motifs of the data sets composed by 4 or less sequences, and of 3 for the other ones (more than 4 sequences). Normally the binding sites are composed by motifs of all or nearly all sequences, and without this constraint is very easy to predict motifs with a high similarity (even 100%) formed, for example, by candidates of only one sequence. Finally, we have applied the complexity concept. The complexity of the candidate motifs should be considered in order to avoid low complexity solutions, for example, the two candidate motifs 'AAAA' and 'AAAA' are very similar, in fact they are identical, but it is not a meaningful motif. The average complexity for a final motif represents the total complexity score for each candidate motif. We calculate the complexity of a candidate motif by using the following expression:
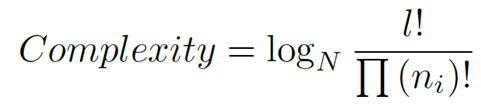
where N = 4 for DNA sequences, l is the motif length, and n_i is the number of nucleotides of type i in {A,C,G,T}. For example, if we consider the motif 'AAAA' (n_A=4, n_T=0, n_G=0, and n_C=0) we will obtain a minimum complexity. Otherwise, if we have, for example, the 'ACGT' motif (n_A=1, n_T=1, n_G=1, and n_C=1) we will obtain a higher complexity. As we can see in the equation, if we do not normalize the complexities when we compare motifs, the maximum complexity was highly dependent of the motif length. The compositional complexity calculation was revised such that the maximum possible complexity score is calculated for each possible motif length prior to the evolutionary computation. During evolution, each complexity score was rescaled between [0,1] where the maximum possible complexity score was 1. This removed any potential bias in complexity relative to the motif length.
Example
Table 1 illustrates an artificial MDP with motif length = 7. By using the motif instances shown in Table 1a and 1c, we obtain the consensus motif A[GT]TTGAA. Due to there is a tie in the second position of the motif, for this position, we select one of the winner nucleotides randomly, in this case we have chosen the nucleotide T. With this consensus motif, in Table 1d we can calculate the value of the second objective. Those sequences whose motif instances exceed a threshold value of concordance of 50% will be taken into account in the support, in this example we have support = 7. The last step is to build the PCM and the PFM by using the nucleotides of the motif instances that have passed the concordance threshold. Having done that, we can obtain the value of the similarity, applying the Similarity equation. In this example we obtain similarity = 0.65.
 |
References
"Predicting DNA Motifs by Using Evolutionary Multiobjective Optimization". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Juan A. Gómez-Pulido, Juan M. Sánchez-Pérez. IEEE Transactions on Systems, Man, and Cybernetics - Part C: Applications and Reviews, Volume 42, Issue 6, 2012, pp. 913-925, ISSN: 1094-6977.
"Comparing Multiobjective Swarm Intelligence Metaheuristics for DNA Motif Discovery". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Juan A. Gómez-Pulido, Juan M. Sánchez-Pérez. Engineering Applications of Artificial Intelligence, Volume 26, Issue 1, Pergamon-Elsevier Science, 2013, pp. 314-326, ISSN: 0952-1976.
"Analysing the Scalability of Multiobjective Evolutionary Algorithms when Solving the Motif Discovery Problem". David L. González-Álvarez, Miguel A. Vega-Rodríguez. Journal of Global Optimization, Volume 57, Issue 2, Springer, 2013, pp. 467-497, ISSN: 0925-5001.
"A Parallel Cooperative Team of Multiobjective Evolutionary Algorithms for Motif Discovery". David L. González-Álvarez, Miguel A. Vega-Rodríguez. Journal of Supercomputing, Volume 66, Issue 3, Springer, 2013, pp. 1576-1612, ISSN: 0920-8542.
"Parallelizing and Optimizing a Hybrid Differential Evolution with Pareto Tournaments for Discovering Motifs in DNA Sequences". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Álvaro Rubio-Largo. Journal of Supercomputing, Volume 70, Issue 2, Springer, 2014, pp. 880-905, ISSN: 0920-8542.
"Gene Variants and Haplotypes Modifying Transcription Factor Binding Sites in the Human Cyclooxygenase 1 and 2 (PTGS1 and PTGS2) Genes". José A.G. Agúndez, David L. González-Álvarez, Miguel A. Vega-Rodríguez, Emilia Botello, Elena García-Martín. Current Drug Metabolism, Volume 15, Issue 2, Bentham Science, 2014, pp. 182-195, ISSN: 1389-2002.
"Convergence Analysis of some Multiobjective Evolutionary Algorithms when Discovering Motifs". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Álvaro Rubio-Largo. Soft Computing, Volume 18, Issue 5, Springer, 2014, pp. 853-869, ISSN: 1432-7643.
"Multiobjective Optimization Algorithms for Motif Discovery in DNA Sequences". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Álvaro Rubio-Largo. Genetic Programming and Evolvable Machines, Volume 16, Issue 2, Springer, 2015, pp. 167-209, ISSN: 1389-2576.
"Finding Patterns in Protein Sequences by Using a Hybrid Multiobjective Teaching Learning Based Optimization Algorithm". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Álvaro Rubio-Largo. IEEE-ACM Transactions on Computational Biology and Bioinformatics, Volume 12, Issue 3, IEEE Computer Society, 2015, pp. 656-666, ISSN: 1545-5963.
"A Comparative Study of Different Motif Occurrence Models applied to a Hybrid Multiobjective Shuffle Frog Leaping Algorithm". David L. González-Álvarez, Miguel A. Vega-Rodríguez, Álvaro Rubio-Largo. Computer Journal, Oxford University Press, 2015, pp. 1-19, ISSN: 0010-4620.
